Stereoinformation in SDF/Mol V3000 Files
How Stereoinformation is stored in in SDF/Mol V3000 files?
✍: Guest
![]() Similar to SDF/Mol V2000, stereoinformation can be stored
in SDF/Mol V3000 files in 3 ways: 0D, 2D, and 3D.
Similar to SDF/Mol V2000, stereoinformation can be stored
in SDF/Mol V3000 files in 3 ways: 0D, 2D, and 3D.
- 0D: Stereoinformation in 0 dimension is defined by parity at atom lines as "CFG=n" options.
- 2D: Stereoinformation in 2 dimension is defined by wedge, hatch or wiggly bond styles at bond lines as "CFG=n" options.
- 3D: Stereoinformation in 3 dimension is defined by the x, y and z 3D coordinates at atom lines.
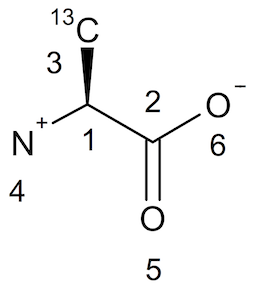
Here is the same molecule L-Alanine with a chiral center (Tetrahedral Stereocenter) stored in a SDF/Mol V3000 file, L-Alanine.sd3:
L-Alanine FYICenter.com Stereoinformation in SDF/Mol V3000 0 0 0 0 0 999 V3000 M V30 BEGIN CTAB M V30 COUNTS 6 5 0 0 1 M V30 BEGIN ATOM M V30 1 C -0.6622 0.5342 0 0 CFG=2 M V30 2 C 0.6622 -0.3 0 0 M V30 3 C -0.7207 2.0817 0 0 MASS=13 M V30 4 N -1.8622 -0.3695 0 0 CHG=1 M V30 5 O 0.622 -1.8037 0 0 M V30 6 O 1.9464 0.4244 0 0 CHG=-1 M V30 END ATOM M V30 BEGIN BOND M V30 1 1 1 2 M V30 2 1 1 3 CFG=1 M V30 3 1 1 4 M V30 4 2 2 5 M V30 5 1 2 6 M V30 END BOND M V30 END CTAB M END
The molecule structure is displayed below:
Open Babel can also be used to read and write SDF/Mol V3000 files.
Here is the command to read a SDF/Mol V3000 file and write in SDF/Mol V2000 format. The conversion keeps both 0D and 2D Stereoinformation.
herong$ obabel -i sdf L-Alanine.sd3 -o sdf
L-Alanine
OpenBabel02032119432D
Stereoinformation in SDF/Mol V3000
6 5 0 0 0 0 0 0 0 0999 V2000
-0.6622 0.5342 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
0.6622 -0.3000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7207 2.0817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8622 -0.3695 0.0000 N 0 3 0 0 0 0 0 0 0 0 0 0
0.6220 -1.8037 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9464 0.4244 0.0000 O 0 5 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
1 3 1 1 0 0 0
1 4 1 0 0 0 0
2 5 2 0 0 0 0
2 6 1 0 0 0 0
M ISO 1 3 13
M CHG 2 4 1 6 -1
M END
$$$$
1 molecule converted
Here is the command to read a SDF/Mol V2000 file and write in SDF/Mol V3000 format.
fyicenter$ obabel L-Alanine.sdf -o sdf -x3 L-Alanine OpenBabel02032111532D 123456789012345678901234567890123456789012345678901234567890 0 0 0 0 0 999 V3000 M V30 BEGIN CTAB M V30 COUNTS 6 5 0 0 1 M V30 BEGIN ATOM M V30 1 C -0.6622 0.5342 0 0 M V30 2 C 0.6622 -0.3 0 0 M V30 3 C -0.7207 2.0817 0 0 MASS=13 M V30 4 N -1.8622 -0.3695 0 0 CHG=1 M V30 5 O 0.622 -1.8037 0 0 M V30 6 O 1.9464 0.4244 0 0 CHG=-1 M V30 END ATOM M V30 BEGIN BOND M V30 1 1 1 2 M V30 2 1 1 3 CFG=1 M V30 3 1 1 4 M V30 4 2 2 5 M V30 5 1 2 6 M V30 END BOND M V30 END CTAB M END $$$$ 1 molecule converted
Open Babel keeps the 2D Stereoinformation correctly. But it fails to added CFG=2 in atom line #1, because it was given in the input SDF/Mol V2000 file:
fyicenter$ cat L-Alanine.sdf
L-Alanine
CTFile Formats, 2003
123456789012345678901234567890123456789012345678901234567890
6 5 0 0 1 0 3 V2000
-0.6622 0.5342 0.0000 C 0 0 2 0 0 0
0.6622 -0.3000 0.0000 C 0 0 0 0 0 0
-0.7207 2.0817 0.0000 C 1 0 0 0 0 0
-1.8622 -0.3695 0.0000 N 0 3 0 0 0 0
0.6220 -1.8037 0.0000 O 0 0 0 0 0 0
1.9464 0.4244 0.0000 O 0 5 0 0 0 0
1 2 1 0 0 0
1 3 1 1 0 0
1 4 1 0 0 0
2 5 2 0 0 0
2 6 1 0 0 0
M CHG 2 4 1 6 -1
M ISO 1 3 13
M END
⇒ Stereochemistry with JSME (Molecule Editor in JavaScript)
⇐ Open Babel Options on SDF/Mol Files
2021-02-06, 3578🔥, 0💬