UI Bug in Embedded 3Dmol Viewer
Why am I not able to load a PDB protein on the Embedded 3Dmol Viewer UI?
✍: FYIcenter.com
![]() The current version of Embedded 3Dmol Viewer has a small code bug.
It labels the dropdown list for data loading incorrectly.
It labels PDB as "cid" and CID as "pdb".
The current version of Embedded 3Dmol Viewer has a small code bug.
It labels the dropdown list for data loading incorrectly.
It labels PDB as "cid" and CID as "pdb".
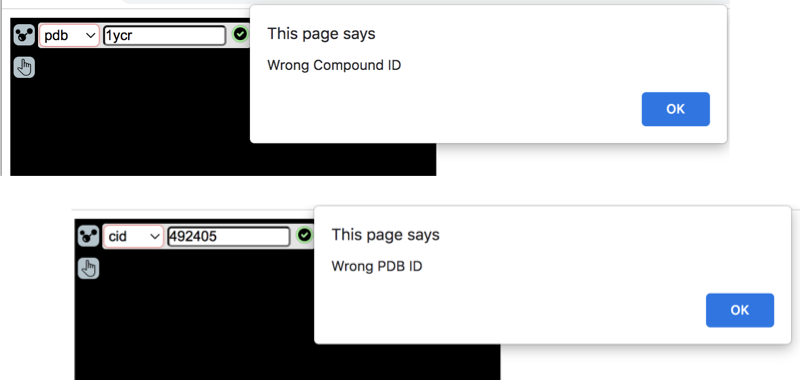
For example, if you select "pdb" and enter "1YCR", you will see the "Wrong Compound ID" error.
If you select "cid" and enter "492405", you will see the "Wrong PDB ID" error.
So you need to select "cid" to load a PDB protein, and select "pdb" to load a CID molecule.
⇒ Play with Embedded 3Dmol Viewer
⇐ UI Components of Embedded 3Dmol Viewer
⇑⇑ 3Dmol.js FAQ
2022-12-26, 1164🔥, 0💬