addModel() - Add Model from Data
How to create a model from a molecule data string with 3Dmol.js addModel() methods?
✍: FYIcenter.com
![]() viewer.addModel() method in the $3Dmol.GLViewer class
allows you to create a model from molecule data string:
viewer.addModel() method in the $3Dmol.GLViewer class
allows you to create a model from molecule data string:
{$3Dmol.GLViewer} addModel(data, format, options) -> {$3Dmol.GLModel}
Here is an HTML code example, View-PDB-Online.html, that create a model from molecule data obtained an AJAX call.
<html><head>
<script src="/3Dmol-min.js"></script>
<style>
.mol-container {width: 400px; height: 400px; position: relative;}
</style>
</head>
<body>
<div id="container-01" class="mol-container"></div>
<script>
$(function() {
let element = $('#container-01');
let config = { backgroundColor: 'grey' };
let viewer = $3Dmol.createViewer( element, config );
let pdbUri = 'https://files.rcsb.org/download/1YCR.pdb';
jQuery.ajax( pdbUri, {
success: function(data) {
let v = viewer;
v.addModel( data, "pdb" ); /* load data */
v.setStyle({}, {cartoon: {color: 'spectrum'}}); /* style all atoms */
v.zoomTo(); /* set camera */
v.render(); /* render scene */
},
error: function(hdr, status, err) {
console.error( "Failed to load PDB " + pdbUri + ": " + err );
},
});
});
</script>
</body></html>
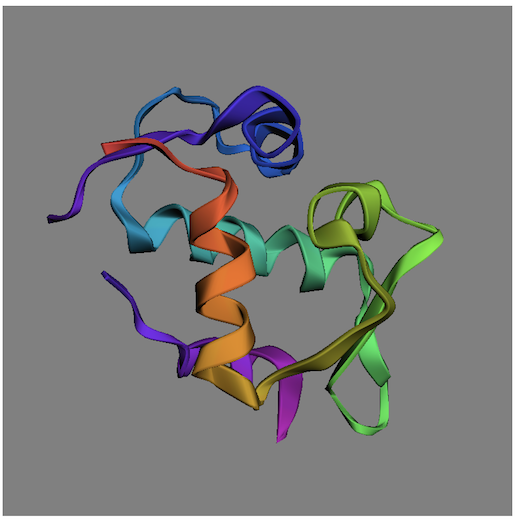
This example HTML code will display a protein structure as shown below:
⇐ addSphere() - Create Spheres
⇑⇑ 3Dmol.js FAQ
2022-12-26, 1166🔥, 0💬