Reaction with Stereocenters in CDXML Format Error
Why am I getting CDXML loader errors on reactions with stereocenters?
✍: FYIcenter.com
![]() Ketcher 2.11.0 has a code bug that gives you a CDXML loader error
if you try to load a reaction with stereocenters in CDXML (ChemDraw XML)
format as shown in this tutorial.
Ketcher 2.11.0 has a code bug that gives you a CDXML loader error
if you try to load a reaction with stereocenters in CDXML (ChemDraw XML)
format as shown in this tutorial.
1. Open Ketcher editor as shown in previous tutorial.
2. Create a dummy reaction with this SMILES string "Br[C@@]([H])(C)O>>Br[C@@](C)([H])O". It has a stereocenter in the reactant and a stereocenter in the product.
3. Select the "Save as" button in the action toolbar at the top of the editor window. You see the "Save Structure" dialog box displayed.
4. Select "CDXML" from the "File Format" dropdown list. You see the reaction is exported in CDXML format:
<?xml version="1.0" encoding="UTF-8"?>
<!DOCTYPE CDXML SYSTEM "http://www.cambridgesoft.com/xml/cdxml.dtd">
<CDXML BondLength="30.000000" LabelFont="3" CaptionFont="4">
<fonttable id="1">
<font id="2" charset="utf-8" name="Arial"/>
<font id="3" charset="utf-8" name="Times New Roman"/>
</fonttable>
<colortable>
<color r="1" g="1" b="1"/>
<color r="0" g="0" b="0"/>
<color r="1" g="0" b="0"/>
<color r="1" g="1" b="0"/>
<color r="0" g="1" b="0"/>
<color r="0" g="1" b="1"/>
<color r="0" g="0" b="1"/>
<color r="1" g="0" b="1"/>
<color r="0.5" g="0.5" b="0.5"/>
</colortable>
<page HeightPages="1" WidthPages="1">
<fragment id="4">
<n id="5" Element="35" NumHydrogens="0" p="122.900780 71.947113">
<t p="122.900780 71.947113" LabelJustification="Left">
<s font="3" size="10" face="96">Br</s>
</t>
</n>
<n id="6" p="122.900780 101.947113" Geometry="Tetrahedral" BondOrdering="7 8 9 5" EnhancedStereoType="Absolute"/>
<n id="7" Element="8" NumHydrogens="1" p="92.900780 101.947113">
<t p="92.900780 101.947113" LabelJustification="Left">
<s font="3" size="10" face="96">OH</s>
</t>
</n>
<n id="8" p="137.900772 127.927879"/>
<n id="9" Element="1" p="151.878540 94.182549">
<t p="151.878540 94.182549" LabelJustification="Left">
<s font="3" size="10" face="96">H</s>
</t>
</n>
<b id="19" B="5" E="6"/>
<b id="20" B="6" E="7"/>
<b id="21" B="6" E="8" Display="WedgeBegin"/>
<b id="22" B="6" E="9" Display="WedgedHashBegin"/>
<graphic BoundingBox="151.878540 71.947113 151.878540 71.947113" GraphicType="Symbol" SymbolType="Absolute" FrameType="None">
<t p="151.878540 71.947113" Justification="Center" InterpretChemically="no">
<s font="3" size="10" face="96">Chiral</s>
</t>
</graphic>
</fragment>
<fragment id="10">
<n id="11" Element="35" NumHydrogens="0" p="297.635254 71.197113">
<t p="297.635254 71.197113" LabelJustification="Left">
<s font="3" size="10" face="96">Br</s>
</t>
</n>
<n id="12" p="297.635254 101.197105" Geometry="Tetrahedral" BondOrdering="15 13 14 11" EnhancedStereoType="Absolute"/>
<n id="13" Element="8" NumHydrogens="1" p="267.635254 101.197105">
<t p="267.635254 101.197105" LabelJustification="Left">
<s font="3" size="10" face="96">OH</s>
</t>
</n>
<n id="14" Element="1" p="312.635254 127.177879">
<t p="312.635254 127.177879" LabelJustification="Left">
<s font="3" size="10" face="96">H</s>
</t>
</n>
<n id="15" p="326.613037 93.432541"/>
<b id="23" B="11" E="12"/>
<b id="24" B="12" E="13"/>
<b id="25" B="12" E="14" Display="WedgeBegin"/>
<b id="26" B="12" E="15" Display="WedgedHashBegin"/>
<graphic BoundingBox="326.613037 71.197113 326.613037 71.197113" GraphicType="Symbol" SymbolType="Absolute" FrameType="None">
<t p="326.613037 71.197113" Justification="Center" InterpretChemically="no">
<s font="3" size="10" face="96">Chiral</s>
</t>
</graphic>
</fragment>
<arrow id="16" BoundingBox="230.379150 99.187500 174.139664 99.187500" FillType="None" ArrowheadType="Solid" HeadSize="2250" ArrowheadWidth="563" ArrowheadHead="Full" ArrowheadCenterSize="25" Head3D="230.379150 99.187500 0.000000" Tail3D="174.139664 99.187500 0.000000"/>
<scheme>
<step ReactionStepReactants="4" ReactionStepProducts="10" ReactionStepArrows="16"/>
</scheme>
</page>
</CDXML>
5. Click the "Save" button to save the structure in a file called ketcher.cdxml.
6. Click the "Open" button in the action toolbar. You see the "Open Structure" dialog box displayed.
7. Click the "OPEN FROM FILE" button to load the structure back from the saved file, ketcher.cdxml. You see an error message displayed:
Convert error! Given string could not be loaded as (query or plain) molecule or reaction, see the error messages: 'CDXML loader: Not a molecule. Found 1 arrows.', 'CDXML loader: stereo type specified for atom #1, but the bond directions does not say that it is a stereocenter', 'CDXML loader: stereo type specified for atom #1, but the bond directions does not say that it is a stereocenter', 'CDXML loader: Queries not supported'
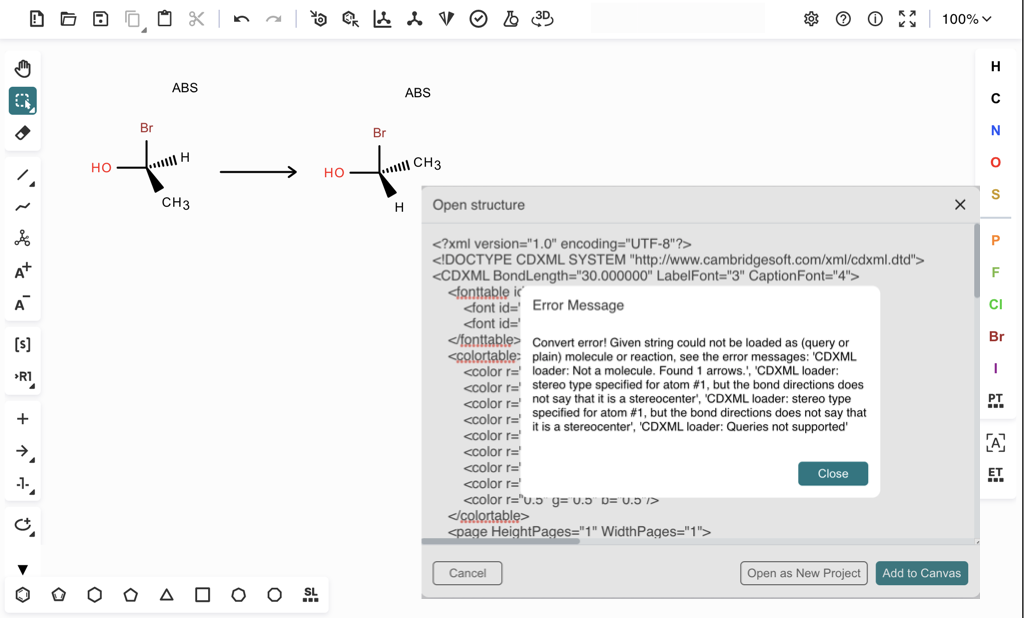
To avoid the loader error, you can modify the CDXML file by removing the EnhancedStereoType="Absolute" attribute from those 2 stereocenters. Then load the modified CDXML file. You see the reaction loaded back in Ketcher correctly.
⇒ Ketcher Chemical Reaction Online Editor
⇐ Show InChI String on Ketcher
2024-01-10, 1115🔥, 0💬