Import Ketcher File to Editor
How to import Ketcher file to the Ketcher editor?
✍: FYIcenter.com
![]() If you created a Ketcher file, you can verify it by importing
it to the Ketcher editor as described below:
If you created a Ketcher file, you can verify it by importing
it to the Ketcher editor as described below:
1. Create a Ketcher file, cyclobutane.ket, with a text editor:
{
"root": {
"nodes": [
{ "$ref": "mol0" }
]
},
"mol0": {
"type": "molecule",
"atoms": [
{
"label": "C",
"location": [ 0, 0, 0 ]
},
{
"label": "C",
"location": [ 1, 0, 0 ]
},
{
"label": "C",
"location": [ 1, 1, 0 ]
},
{
"label": "C",
"location": [ 0, 1, 0 ]
}
],
"bonds": [
{
"type": 1,
"atoms": [ 0, 1 ]
},
{
"type": 1,
"atoms": [ 1, 2 ]
},
{
"type": 1,
"atoms": [ 2, 3 ]
},
{
"type": 1,
"atoms": [ 3, 0 ]
}
]
}
}
2. Open Ketcher editor as shown in previous tutorials.
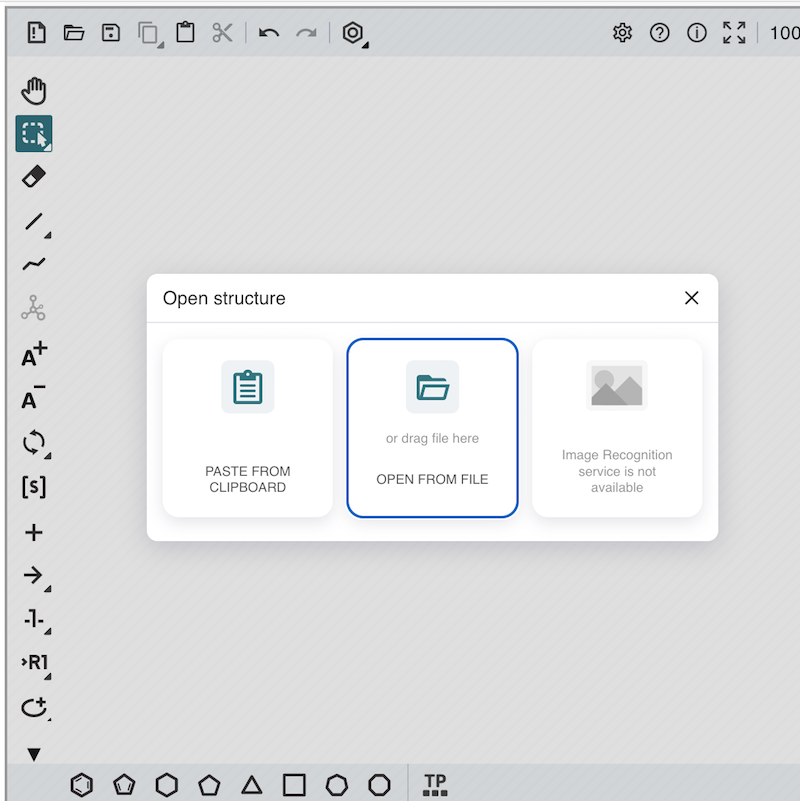
3. Click the "open" icon in the action toolbar at the top of the editor window. You see the "Open Structure" dialog box displayed.
4. Click the "OPEN FROM FILE" button to import the Ketcher file, cyclobutane.ket. You see a cyclobutane molecule structure displayed in the editor.
⇒ Ketcher File Structure for Molecule
⇐ Export Ketcher File from Editor
2024-03-23, 1036🔥, 0💬