Build Molecule Pattern in JSME
How to build a molecule pattern in JSME editor? I also want to save it a SMARTS string to do substructure search.
✍: FYIcenter.com
![]() If you want to build a molecule pattern in JSME, you can follow this tutorial.
If you want to build a molecule pattern in JSME, you can follow this tutorial.
1. Follow the previous tutorial to install JSME on your Web server.
2. Run the demo page on the Apache Web server by opening "http://localhost/jsme/JSME.html" URL in a Web browser.
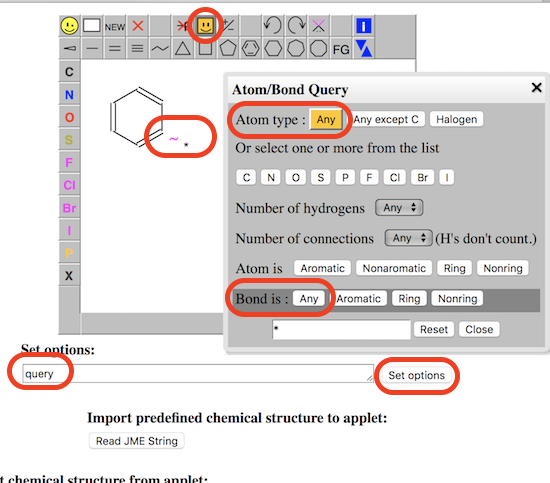
3. Enter "query" in the options input box below the editor and click "Set options". You see the "SMARTS" icon displayed in the menu.
4. Select the "benzene ring" icon from the JSME editor menu and click a location inside the editor. You see a benzene ring molecule created.
5. Select the "SMARTS" icon from the menu. and click "Bond is [Any]". Then add an "Any" bond to the benzene ring.
6. Select the "SMARTS" icon from the menu. and click "Atom type: [Any]". Then add an "Any" atom to the end of the "any" bond.
7. Select the "export & import" icon from the menu and select "Copy as SMILES". You see the SMARTS string "C/1(~*)=C/C=C\C=C1" displayed.
⇐ Show InChI and InChIKey on JSME
2020-05-18, 2678🔥, 0💬