Substructure Search with SMARTS Expressions
How to use SMARTS expressions in a substructure search using "babel" commands?
✍: FYIcenter.com
![]() You can use SMARTS expressions in the "-s ..." option in
"babel" commands to filter molecules that match given SMARTS expressions.
You can use SMARTS expressions in the "-s ..." option in
"babel" commands to filter molecules that match given SMARTS expressions.
Here are some examples:
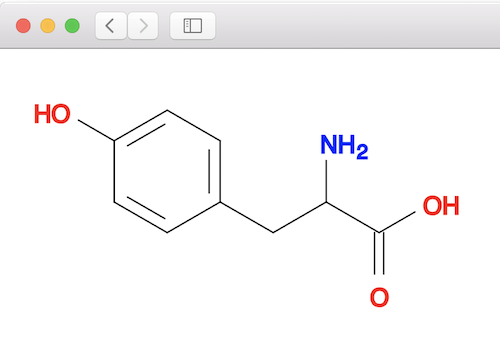
# C, C and O connected with single bonds fyicenter$ obabel "-:c1cc(ccc1CC(C(=O)O)N)O" -o smiles -s C-C-O c1cc(ccc1CC(C(=O)O)N)O 1 molecule converted # same as above with single bonds omitted fyicenter$ obabel "-:c1cc(ccc1CC(C(=O)O)N)O" -o smiles -s CCO c1cc(ccc1CC(C(=O)O)N)O 1 molecule converted # same as above atom expression with optional brackets added fyicenter$ obabel "-:c1cc(ccc1CC(C(=O)O)N)O" -o smiles -s C[C]O c1cc(ccc1CC(C(=O)O)N)O 1 molecule converted # double conditions on the middle atom fyicenter$ obabel "-:c1cc(ccc1CC(C(=O)O)N)O" -o smiles -s C[CH0]O c1cc(ccc1CC(C(=O)O)N)O 1 molecule converted # same as above with implicit & included fyicenter$ obabel "-:c1cc(ccc1CC(C(=O)O)N)O" -o smiles -s 'C[C&H0]O' c1cc(ccc1CC(C(=O)O)N)O 1 molecule converted # bond expression used fyicenter$ obabel "-:c1cc(ccc1CC(C(=O)O)N)O" -o smiles -s 'C-,=O' c1cc(ccc1CC(C(=O)O)N)O 1 molecule converted # same as above, but in a native way fyicenter$ obabel "-:c1cc(ccc1CC(C(=O)O)N)O" -o smiles -s 'C!#O' c1cc(ccc1CC(C(=O)O)N)O 1 molecule converted # bad bond expression, no bond can be both single and double. fyicenter$ obabel "-:c1cc(ccc1CC(C(=O)O)N)O" -o smiles -s 'C-=O' 0 molecules converted # poor bond expression, a single bond is also an any bond. fyicenter$ obabel "-:c1cc(ccc1CC(C(=O)O)N)O" -o smiles -s 'C-~O' c1cc(ccc1CC(C(=O)O)N)O 1 molecule converted # matching aromatic C and connected with 1 H fyicenter$ obabel "-:c1cc(ccc1CC(C(=O)O)N)O" -o smiles -s '[c;H1]' c1cc(ccc1CC(C(=O)O)N)O 1 molecule converted # matching aromatic C and connected with 0 H fyicenter$ obabel "-:c1cc(ccc1CC(C(=O)O)N)O" -o smiles -s '[c;H0]' c1cc(ccc1CC(C(=O)O)N)O 1 molecule converted # nested SMARTS expressions fyicenter$ obabel "-:c1cc(ccc1CC(C(=O)O)N)O" -o smiles -s '[C;H0]-,=[$([O;H1]),$([O;H0])]' c1cc(ccc1CC(C(=O)O)N)O 1 molecule converted
You can validate the above matching result by looking at the tyrosine molecule structure below:
⇒ Similarity Search with Open Babel
2021-01-09, 1788🔥, 0💬