"obgen" - Generate Molecule 3D Structures
What Is "obgen" command? How to use it to Generate Molecule 3D Structures?
✍: FYIcenter.com
![]() "obgen" command is a command line tool provided in the Open Babel package
that allows you to Generate Molecule 3D Structures.
"obgen" command does the same job as the "babel ... --gen3D" with more options.
"obgen" command is a command line tool provided in the Open Babel package
that allows you to Generate Molecule 3D Structures.
"obgen" command does the same job as the "babel ... --gen3D" with more options.
Here is the user manual of the "obgen" command.
NAME
obgen -- generate 3D coordinates for a molecule
SYNOPSIS
obgen [OPTIONS] filename
DESCRIPTION
The obgen tool will generate 3D coordinates for molecules in a file (e.g.
multi-molecule SMILES files). The resulting structure will be optimized
using the given forcefield and checked for the lowest-energy conformer
using a Monte Carlo search. Output will be sent to standard output in
the SDF file format.
OPTIONS
If no filename is given, obgen will give all options including the available
forcefields.
-ff forcefield
Select the forcefield
EXAMPLES
View the possible options, including available forcefields:
obgen
Generate 3D coordinates for the molecule(s) in file test.smi:
obgen test.smi
Generate 3D coordinates for the molecule(s) in file test.smi using the UFF forcefield:
obgen -ff UFF test.smi
Here is an example of generating, or converting, a given molecule structure to the best conforming 3D structure using the default force field type:
1. Create a 2D structure of the tyrosine molecule:
fyicenter$ obabel "-:c1cc(ccc1CC(C(=O)O)N)O" -O tyrosine.sdf --gen2D 1 molecule converted fyicenter$ babel tyrosine.sdf tyrosine.svg 1 molecule converted 29 audit log messages
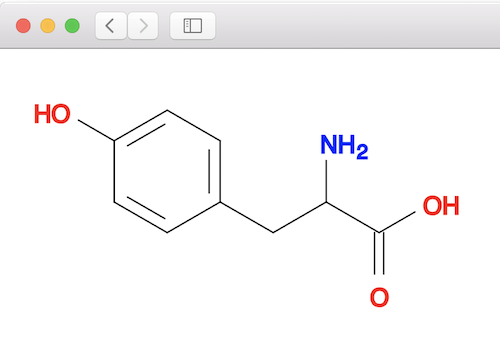
2. Generate, or convert, the 2D structure to 3D structure using the "obgen" command:
fyicenter$ obgen tyrosine.sdf > tyrosine-3d-obgen.sdf
A T O M T Y P E S
IDX TYPE
1 37
2 37
3 37
...
F O R M A L C H A R G E S
IDX CHARGE
1 0.000000
2 0.000000
3 0.000000
...
P A R T I A L C H A R G E S
IDX CHARGE
1 -0.150000
2 -0.150000
3 0.082500
...
S E T T I N G U P C A L C U L A T I O N S
SETTING UP BOND CALCULATIONS...
SETTING UP ANGLE & STRETCH-BEND CALCULATIONS...
SETTING UP TORSION CALCULATIONS...
SETTING UP OOP CALCULATIONS...
SETTING UP VAN DER WAALS CALCULATIONS...
SETTING UP ELECTROSTATIC CALCULATIONS...
S T E E P E S T D E S C E N T
STEPS = 500
STEP n E(n) E(n-1)
------------------------------------
0 124.930 ----
10 43.09167 44.83204
20 29.55319 29.87195
30 20.14396 22.92951
...
STEEPEST DESCENT HAS CONVERGED
W E I G H T E D R O T O R S E A R C H
NUMBER OF ROTATABLE BONDS: 3
NUMBER OF POSSIBLE ROTAMERS: 288
INITIAL WEIGHTING OF ROTAMERS...
GENERATED 250 CONFORMERS
CONFORMER ENERGY
--------------------
1 11.188
2 4.809
3 3.506
.. ...
248 6.265
249 11.152
250 9.037
LOWEST ENERGY: 0.926
S T E E P E S T D E S C E N T
STEPS = 500
STEP n E(n) E(n-1)
------------------------------------
0 0.926 ----
10 0.92061 0.92084
20 0.91410 0.91561
30 0.90976 0.91002
...
STEEPEST DESCENT HAS CONVERGED
fyicenter$ babel tyrosine-3d-obgen.sdf tyrosine-3d-obgen.svg -d
1 molecule converted
34 audit log messages
You need a 3D structure viewer to see the 3D structure. But you can look at the 2D presentation of the 3D structure.
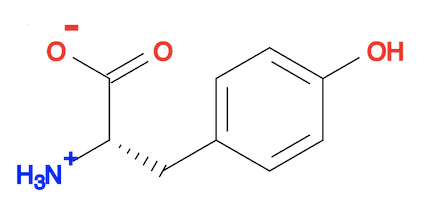
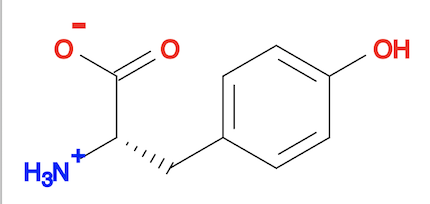
3. You can actually use "babel ... --gen3D" command to do same job as the "obgen" command. The result is the same as the "obgen" command.
fyicenter$ babel tyrosine.sdf tyrosine-3d-babel.sdf --gen3D 1 molecule converted 118 audit log messages 3 debugging messages fyicenter$ babel tyrosine-3d-babel.sdf tyrosine-3d-babel.svg -d 1 molecule converted 34 audit log messages
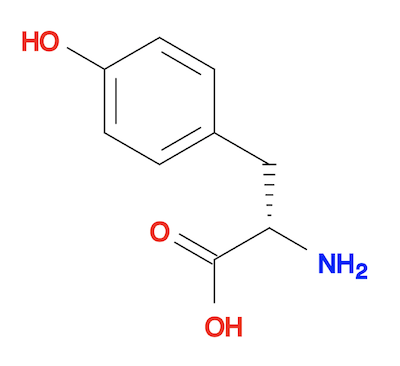
4. Or you can generate the 3D structure from the SMILES string directly using the "obabel" command. The result is almost the same as the "obgen" command.
fyicenter$ obabel "-:c1cc(ccc1CC(C(=O)O)N)O" -O tyrosine-3d-def.sdf --gen3D 1 molecule converted fyicenter$ babel tyrosine-3d-def.sdf tyrosine-3d-def.svg -d 1 molecule converted 34 audit log messages
⇒ "obgrep" - Search Molecules using SMARTS
2020-11-11, 3999🔥, 0💬