"obminimize" - Optimize Geometry/Energy of Molecule
What Is "obminimize" command? How to use it to optimize the geometry and minimize the energy for a given molecule?
✍: FYIcenter.com
![]() "obminimize" command is a command line tool provided in the Open Babel package
that allows you to optimize the geometry and minimize the energy for a given molecule.
"obminimize" command is a command line tool provided in the Open Babel package
that allows you to optimize the geometry and minimize the energy for a given molecule.
Here is the user manual of the "obminimize" command.
NAME
obminimize -- optimize the geometry, minimize the energy for a molecule
SYNOPSIS
obminimize [OPTIONS] filename
DESCRIPTION
The obminimize tool can be used to minimize the energy for molecules inside (multi-)molecule files (e.g., MOL2,
etc.)
OPTIONS
If no filename is given, obminimize will give all options including the available forcefields.
-n steps
Specify the maximum number of steps (default=2500)
-cg Use conjugate gradients algorithm (default)
-sd Use steepest descent algorithm
-c criteria
Set convergence criteria (default=1e-6)
-ff forcefield
Select the forcefield
EXAMPLES
View the possible options, including available forcefields:
obminimize
Minimize the energy for the molecule(s) in file test.mol2:
obminimize test.mol2
Minimize the energy for the molecule(s) in file test.mol2 using the Ghemical
forcefield:
obminimize -ff Ghemical test.mol2
Minimize the energy for the molecule(s) in file test.mol2 by taking at most
300 geometry optimization steps
obminimize -n 300 test.mol2
Minimize the energy for the molecule(s) in file test.mol2 using the steepest
descent algorithm and convergence criteria 1e-5:
obminimize -sd -c 1e-5 test.mol2
Here is an example of minimizing the tyrosine molecule with the "obminimize" command:
1. Create a 2D structure of the tyrosine molecule:
fyicenter$ obabel "-:c1cc(ccc1CC(C(=O)O)N)O" -O tyrosine.sdf --gen2D 1 molecule converted fyicenter$ babel tyrosine.sdf tyrosine.svg 1 molecule converted 29 audit log messages
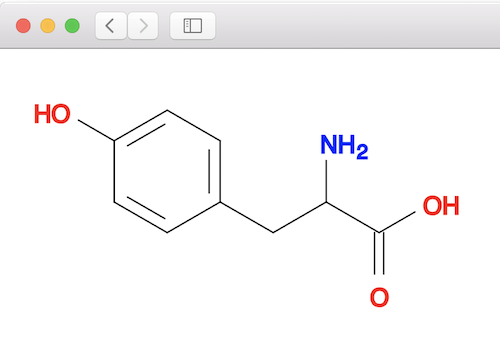
2. Optimize the geometry and minimize the energy of the tyrosine molecule. Note that "obminimize" command creates the output molecule in PDB format.
fyicenter$ obminimize tyrosine.sdf > tyrosine-minimized.pdb
A T O M T Y P E S
IDX TYPE
1 0603
2 0603
3 0603
4 0603
5 0603
6 0603
7 0600
8 0600
9 0601
10 0801
11 0800
12 0700
13 0800
C H A R G E S
IDX CHARGE
1 0.000000
2 0.000000
3 0.000000
4 0.000000
5 0.000000
6 0.000000
7 0.000000
8 0.000000
9 0.100000
10 -0.100000
11 0.000000
12 0.000000
13 0.000000
S E T T I N G U P C A L C U L A T I O N S
SETTING UP BOND CALCULATIONS...
SETTING UP ANGLE CALCULATIONS...
SETTING UP TORSION CALCULATIONS...
SETTING UP VAN DER WAALS CALCULATIONS...
SETTING UP ELECTROSTATIC CALCULATIONS...
C O N J U G A T E G R A D I E N T S
STEPS = 2500
STEP n E(n) E(n-1)
--------------------------------
1 10225.625 10903.871
2 7582.300 7582.300
CONJUGATE GRADIENTS HAS CONVERGED
Time: 0seconds. Iterations per second: inf
3. See what the minimized molecule structure looks like:
fyicenter$ babel tyrosine-minimized.pdb tyrosine-minimized.svg 1 molecule converted 28 audit log messages
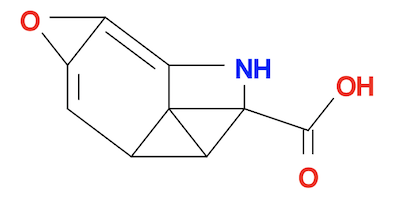
⇒ "obprobe" - Create Electrostatic Probe Grid
2022-08-27, 9091🔥, 2💬