$3Dmol.download('pdb:...', ...) - Download PDB Protein
How to download PDB Protein Data with the $3Dmol.download() method?
✍: FYIcenter.com
![]() The syntax to download PDB Protein Data with the $3Dmol.download() method
is shown below:
The syntax to download PDB Protein Data with the $3Dmol.download() method
is shown below:
$3Dmol.download("pdb:{PDB_ID}", viewer, options, callback) -> {$3Dmol.GLModel}
viewer - The $3Dmol.GLViewer instance where the downloaded protein is added
options - Options applied to the downloaded data
callback - Function to call after data is loaded with model as argument
Here is an HTML code example, Download-PDB.html, that uses the download() method to download a protein with a given PDB ID.
<html><head>
<script src="/3Dmol-min.js"></script>
</head>
<body>
<div id="viewer" style="width: 400px; height: 400px; position: relative;"></div>
<script>
let config = { backgroundColor:'grey' };
let viewer = $3Dmol.createViewer( $('#viewer'), config );
$3Dmol.download("pdb:1MO8", viewer, {}, function(m) {
m.setStyle({"chain":"A"}, {cartoon:{color:"spectrum"}});
viewer.render();
});
</script>
</body></html>
This example HTML code will display a PDB protein in cartoon style:
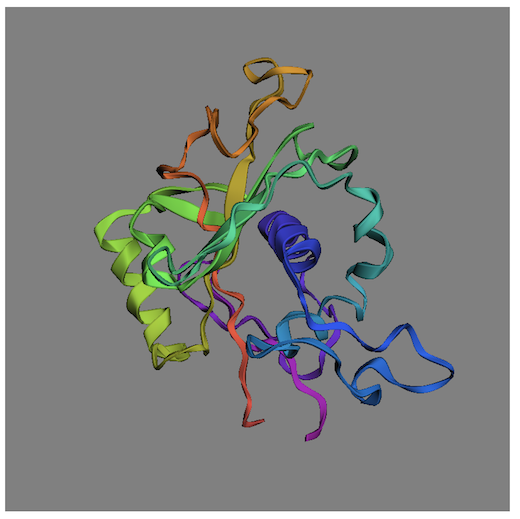
Note that the protein structure data is downloaded with the following PDB API:
http://files.rcsb.org/view/{PDB_ID}.pdb
⇒ $3Dmol.download('cid:...', ...) - Download CID Molecule
⇐ $3Dmol Namespace and Static Methods
⇑⇑ 3Dmol.js FAQ
2023-01-24, 1400🔥, 0💬