"cid={PubChem_CID}" - Load Molecule from PubMed
How to load a molecule structure with the CID in the Online 3Dmol Viewer?
✍: FYIcenter.com
![]() You can use the cid={PubChem_CID} URL parameter to load a molecule structure from
https://pubchem.ncbi.nlm.nih.gov/ in the Online 3Dmol Viewer.
You can use the cid={PubChem_CID} URL parameter to load a molecule structure from
https://pubchem.ncbi.nlm.nih.gov/ in the Online 3Dmol Viewer.
https://3dmol.org/viewer.html?cid={PubChem_CID}
For example, enter the following URL in Web browser. You see the molecule structure of Aspirin is displayed in the 3Dmol Viewer.
https://3dmol.org/viewer.html?cid=2244&style=stick
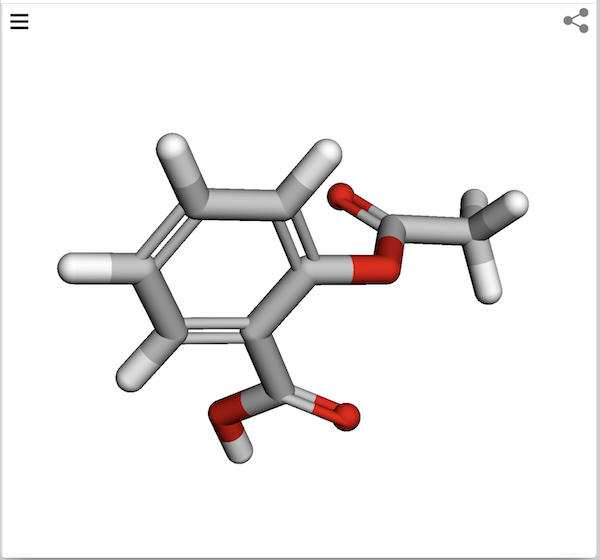
Note that the molecule structure data is loaded with the following PubMed API:
https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/{CID}/SDF?record_type=3d
⇒ "url={URL}" - Load Molecule from URL
⇐ "labelres={...}" - residue Label Properties
⇑ Using Online Server of 3Dmol Viewer
⇑⇑ 3Dmol.js FAQ
2023-09-07, 1422🔥, 0💬