Load Molecule Strcuture from File
How to load a molecule structure from a file into PyMol?
✍: FYIcenter.com
![]() In order to visualize a molecule in PyMol, you need to load
the molecule structure from a file first.
In order to visualize a molecule in PyMol, you need to load
the molecule structure from a file first.
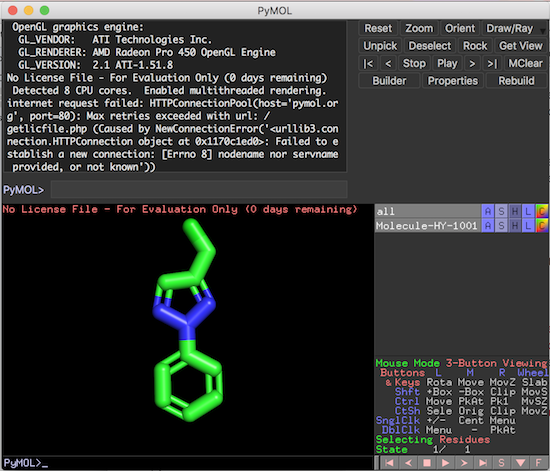
1. Start PyMol and click the "File > Open" menu. You see the file open dialog box.
2. Find and select the "Molecule-HY-001.sdf" file, which stores a molecule structure in a SDF (Structure Data File) file.
3. Click "Open" to load the molecule structure from the file into PyMol. You see the molecule structure is displayed in the viewer area.
Now you can use PyMol commands or mouse clicks to control how the molecule structure is displayed.
Â
⇒ PyMol Tutorials
⇑⇑ PyMol Tutorials
2020-04-16, 1820🔥, 0💬