Save Molecule from JSME to Server
How to save molecules created in JSME editor to the Web server?
✍: FYIcenter.com
![]() If you want to save the molecule structure created in the JSME editor
to the backend Web server, you need write JavaScript code to extract the
molecule data and send it as HTML form parameters.
If you want to save the molecule structure created in the JSME editor
to the backend Web server, you need write JavaScript code to extract the
molecule data and send it as HTML form parameters.
1. Create a new HTML document, save-molecule.html, to present the JSME editor and the form:
<html>
<!-- save-molecule.html Copyright (c) FYIcenter.com. All rights reserved. -->
<head>
<title>Create and Save Molecule</title>
<script type="text/javascript" src="/jsme/jsme.nocache.js"></script>
<script>
function jsmeOnLoad() {
jsmeObj = new JSApplet.JSME("jsme_container", "380px", "340px");
}
function convert() {
var smiles = jsmeObj.smiles();
var sdf = jsmeObj.molFile();
var svg = jsmeObj.getMolecularAreaGraphicsString();
document.getElementsByName("smiles")[0].value = smiles;
document.getElementsByName("sdf")[0].value = sdf;
document.getElementsByName("svg")[0].value = svg;
updateInChI(sdf);
}
</script>
</head>
<body>
<script>
var JSApplet = {};
JSApplet.Inchi = {};
</script>
<script type="text/javascript" src="/jsme/96E40B969193BD74B8A621486920E79C.cache.js"></script>
<script>
function updateInChI(sdf) {
var result;
JSApplet.Inchi.__resultHandler = function (inchi_result) {
result = inchi_result
};
JSApplet.Inchi.computeInchi(sdf, "JSApplet.Inchi.__resultHandler");
delete JSApplet.Inchi.__resultHandler;
var text = "";
Object.keys(result).forEach(function (k) {
text += k + ": " + result[k] + "\n"
});
document.getElementsByName("inchi")[0].value = text;
}
</script>
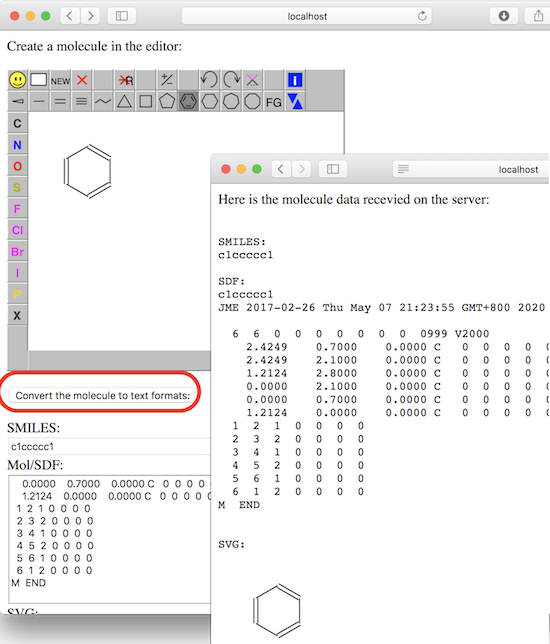
<p>Create a molecule in the editor:</p>
<div id="jsme_container"></div>
<p><button type="button" onclick="convert()">
Convert the molecule to text formats:</button>
</p>
<form action="save-molecule.php" method="post">
SMILES:<br/>
<input type="text" name="smiles" value="" size="62"/><br/>
Mol/SDF:<br/>
<textarea name="sdf" rows="10" cols="60"></textarea><br/>
SVG:<br/>
<textarea name="svg" rows="10" cols="60"></textarea><br/>
InChI and InChIKey:<br/>
<textarea name="inchi" rows="10" cols="60"></textarea><br/>
<input type="submit" value="Save"/>
</form>
</body>
</html>
2. Create PHP script, save-molecule.php, to process data submitted from the form.
<html>
<!-- save-molecule.php Copyright (c) FYIcenter.com. All rights reserved. -->
<head>
<title>Molecule Data Received</title>
</head>
<body>
<p>Here is the molecule data received on the server:
</p>
<pre>
<?php
$smiles = $_REQUEST["smiles"];
$sdf = $_REQUEST["sdf"];
$svg = $_REQUEST["svg"];
$inchi = $_REQUEST["inchi"];
echo("\nSMILES:\n$smiles\n");
echo("\nSDF:\n$sdf\n");
echo("\nSVG:\n$svg\n");
echo("\nInChI and InChIKey:\n$inchi\n");
?>
</pre>
</body>
</html>
3. Copy both save-molecule.html and save-molecule.php to the Web server:
fyicenter$ sudo cp save-molecule.* /var/www/html/jsme
4. Test JSME editor page on the Apache Web server by opening "http://localhost/jsme/save-molecule.html" URL in a Web browser.
5. Select the "benzene ring" icon from the JSME editor menu and click a location inside the editor. You see a benzene ring molecule created.
6. Click "Convert" button below the editor. You see molecule's SMILES string, Mol/SDF and SVG representations populated in the form.
7. Click "Save" button below the form. You see molecule's SMILES string, Mol/SDF and SVG representations received on the server and echoed back to the browser.
⇒ Refresh Molecule in JSME Editor
2020-05-18, 2701🔥, 0💬