Stereochemistry with JSME (Molecule Editor in JavaScript)
Can JSME (Molecule Editor in JavaScript) be used to do Stereochemistry?
✍: Guest
![]() Yes. JSME (Molecule Editor in JavaScript) can be used to stereochemistry.
You can create a molecule structure with
wedges and hatches provide stereoinformation on bonds.
Yes. JSME (Molecule Editor in JavaScript) can be used to stereochemistry.
You can create a molecule structure with
wedges and hatches provide stereoinformation on bonds.
Here is an example with 1 wedge and 1 hatch:
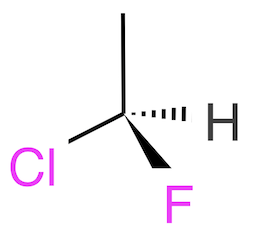
You can then write the structure into a SDF/Mol V2000 file, FYI-002.sdf. The wedge is stored as Stereo code "1" and the hatch is stored as Stereo code "6". In other words, only 2D stereoinformation is recorded.
[H][C@](C)(F)Cl
JME 2017-02-26 Wed Feb 03 20:27:39 GMT+800 2021
5 4 0 0 0 0 0 0 0 0999 V2000
1.2481 1.1538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0000 0.5195 0.0000 Cl 0 0 0 0 0 0 0 0 0 0 0 0
2.6481 1.1469 0.0000 H 0 0 0 0 0 0 0 0 0 0 0 0
2.0411 0.0000 0.0000 F 0 0 0 0 0 0 0 0 0 0 0 0
1.2376 2.5537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
1 3 1 6 0 0 0
1 4 1 1 0 0 0
1 5 1 0 0 0 0
M END
You can also write the structure into a SDF/Mol V3000 file, FYI-002.sd3. Note that "CFG=3" in V3000 is the same as Stereo Code of "6" in V2000 format.
[H][C@](C)(F)Cl JME 2017-02-26 Wed Feb 03 20:29:08 GMT+800 2021 0 0 0 0 0 0 0 0 0 0999 V3000 M V30 BEGIN CTAB M V30 COUNTS 5 4 0 0 0 M V30 BEGIN ATOM M V30 1 C 1.2481 1.1538 0.0000 0 M V30 2 Cl 0.0000 0.5195 0.0000 0 M V30 3 H 2.6481 1.1469 0.0000 0 M V30 4 F 2.0411 0.0000 0.0000 0 M V30 5 C 1.2376 2.5537 0.0000 0 M V30 END ATOM M V30 BEGIN BOND M V30 1 1 1 2 M V30 2 1 1 3 CFG=3 M V30 3 1 1 4 CFG=1 M V30 4 1 1 5 M V30 END BOND M V30 END CTAB M END
JSME can also read stereoinformation from SDF/Mol files.
⇐ Stereoinformation in SDF/Mol V3000 Files
2021-02-06, 1445🔥, 0💬