"select=..." - Select Substructure in 3Dmol Viewer
How to select substructure in the Online 3Dmol Viewer?
✍: FYIcenter.com
![]() You can use the select={Selection} URL parameter to
select a substructure in the loaded molecule and specify a different style
than the whole molecule.
You can use the select={Selection} URL parameter to
select a substructure in the loaded molecule and specify a different style
than the whole molecule.
https://3dmol.org/viewer.html?select={Selection}&style={Style}&select={Selection}&style={Style}...
Each {Selection} can use the following syntax:
select={key:value;key:value;...}
There are 2 types of "key"s supported in the online viewer:
1. Non-AtomSpec selection keys:
all - Select the entire molecule. This is the default.
invert:{true|false} - If "true", inverts the meaning of the selection.
expand:{number} - Expands the selection to include all atoms within a given distance.
byres:{true|false} - If "true", include any residues containing selected atoms.
...
2. AtomSpec selection keys:
resi:{number} - Residue number for protein molecules. For example: resi:100.
resn:{name} - Residue name for protein molecules. For example: resn:TYR.
chain:{letter} - Chain letter for PDB data files. For example: chain:B.
...
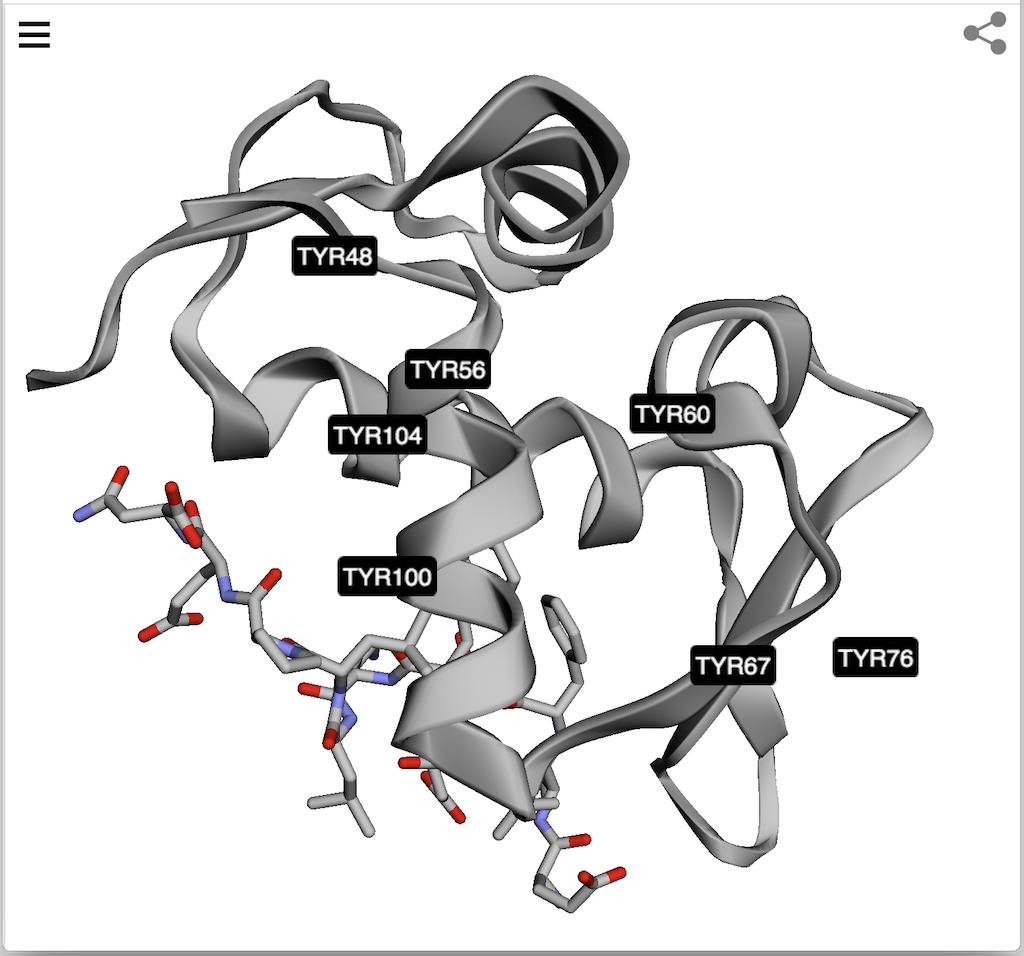
For example, enter the following URL in Web browser.
https://3dmol.org/viewer.html?pdb=1YCR&style=cartoon&select=chain:B&style=stick&select=resn:TYR&labelres=
The loaded PDB file will be displayed with the following selections and their styles:
1. "style=cartoon" - Display the entire molecule in "cartoon" style.
2. "select=chain:B&style=stick" - Display the chain B in "stick" style.
3. "select=resn:TYR&labelres=" - Display the TYR residues with residue labels.
⇒ "select=resi:..." - Select Protein Residues by Numbers
⇐ "style=..." - Specify Display Style in 3Dmol Viewer
⇑ Using Online Server of 3Dmol Viewer
⇑⇑ 3Dmol.js FAQ
2023-09-07, 1091🔥, 0💬